Microbes are the best chemists on Earth
Elucidating the chemistry encoded in human gut bacteriophages
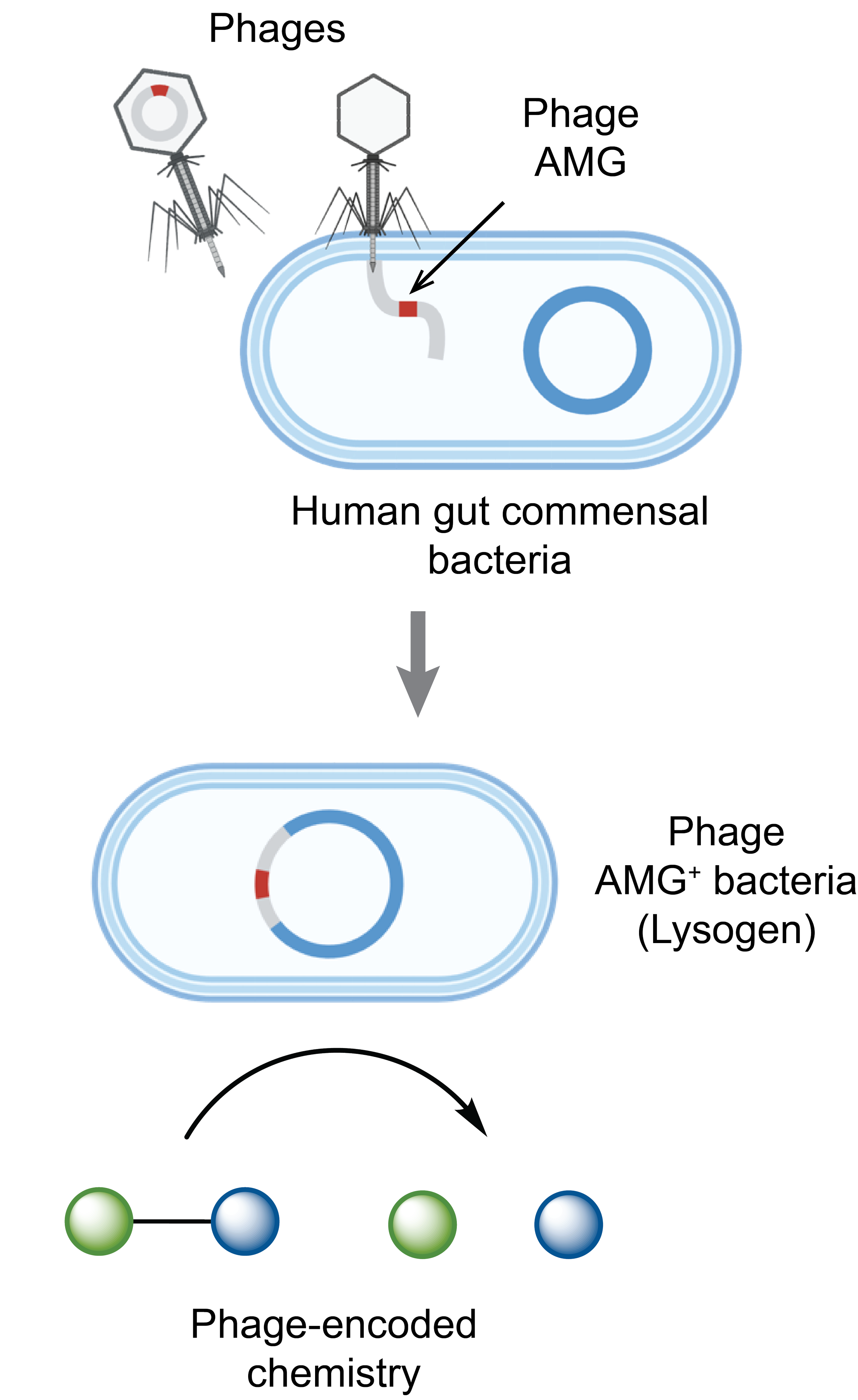
The human gut is home to trillions of microbes, with bacteriophages being the second most numerous entities in the gut microbiome. Bacteriophages, or phages, are obligate parasites that alter the genomes of their bacterial host by introducing their viral genomes, which encode auxiliary metabolic genes (AMGs). However, the ability of phages to expand the chemical repertoire of their bacterial hosts, and the effect of phages on the biochemical dynamics of the gut microbiome are largely unknown. We are investigating the chemistry encoded by human gut phages and its effects on human gut microbial ecology. We use interdisciplinary approaches to uncover the chemistry performed by phage AMGs and elucidate how phage-encoded metabolism affects bacterial and human biology. This platform starts by searching for and characterizing AMGs from phage genomes and transcriptomes using chemical knowledge, links AMG chemistry to phage biology, and assesses the effect of phage chemistry on human health. These studies (i) provide a mechanistic understanding of phage AMG function that may reveal new chemistry, (ii) interrogate the impact of gut phage on bacterial and human biology, and (iii) create new microbiome engineering strategies.
Natural product discovery and chemical ecology in the human oral microbiome
The human oral cavity microbiome represents a rich and untapped source of secondary metabolites that may play important roles in human oral health and disease. However, gaps in our knowledge of the identity of these metabolites and the microbes that produce them hinder our understanding of their causal roles in diseases. We are focused on investigating natural product biosynthesis in the human oral microbiome and establishing the causal link between oral microbial natural products and human oral disease. We are addressing the open questions regarding the biosynthetic gene clusters encoded by cultivated and uncultivated oral bacteria, and the metabolites they produce using bioinformatics and functional characterization. We are using activity-guided fractionation approaches to discover new oral microbial natural products. We then assess the bioactivity of these natural products against oral commensal and pathogenic bacteria. The causal role of these metabolites in human oral health will be established collaboratively using gnotobiotic animal models. Our work will ultimately elucidate the microbial dark matter of the human oral microbiome, and how oral bacterial natural products affect host health and shape oral microbial communities.
Engineering microbiomes for the biodegradation of environmental pollutants
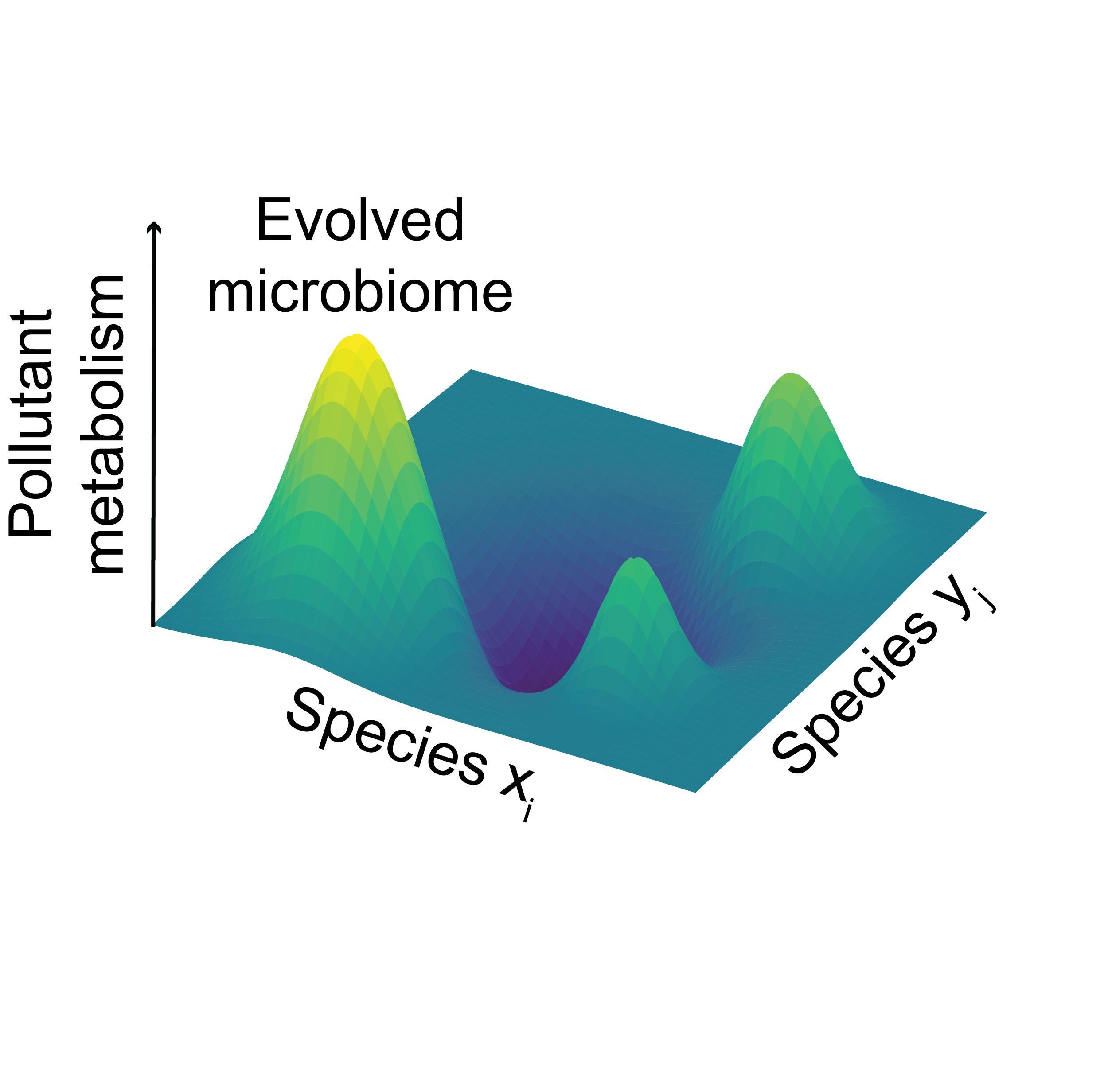
Microbes inhabiting contaminated environmental sites evolve over time to perform chemistry on abiological contaminant substrates. This chemistry is of great interest for the biodegradation of pollutants that pose serious health hazards to humans. Several microbes have recently evolved to metabolize several serious pollutants, albeit with biochemical activities too low to be useful in bioremediation. A strategy that Nature has developed to enable the sharing of beneficial genes among microbial community members, and increase the shared community function is horizontal gene transfer. We are developing directed evolution platforms to evolve environmental microbiomes for the efficient biodegradation of environmental pollutants. To accomplish this goal, we will collect environmental microbial samples, and measure their metabolism of pollutants to prioritize microbial communities for directed evolution. We apply a sexual directed evolution strategy to pollutant-metabolizing microbiomes that combines enrichment culturing, horizontal gene transfer, and systematic biochemical characterization. We will determine the microbial, genetic and molecular determinants for engineered microbial metabolism of pollutants. Our work will ultimately increase our basic understanding of how microbes and microbiomes evolve to catalyze new chemical transformations, and will create new workflows to engineer the metabolic functions of microbial communities.
The Tinoco Lab is a member of the following research centers and institutes:
The Tinoco Lab at UCLA is generously funded by: